Researchers at Arc Institute have created the first AI-generated bacteriophage genomes using genomic foundation models, overcoming evolutionary constraints to produce viable synthetic organisms. This breakthrough enables engineered phages that combat antibiotic-resistant bacteria and showcases AI's ability to design complex biological systems from scratch. The work marks a historic leap from merely reading and writing DNA to actively designing functional genomes.
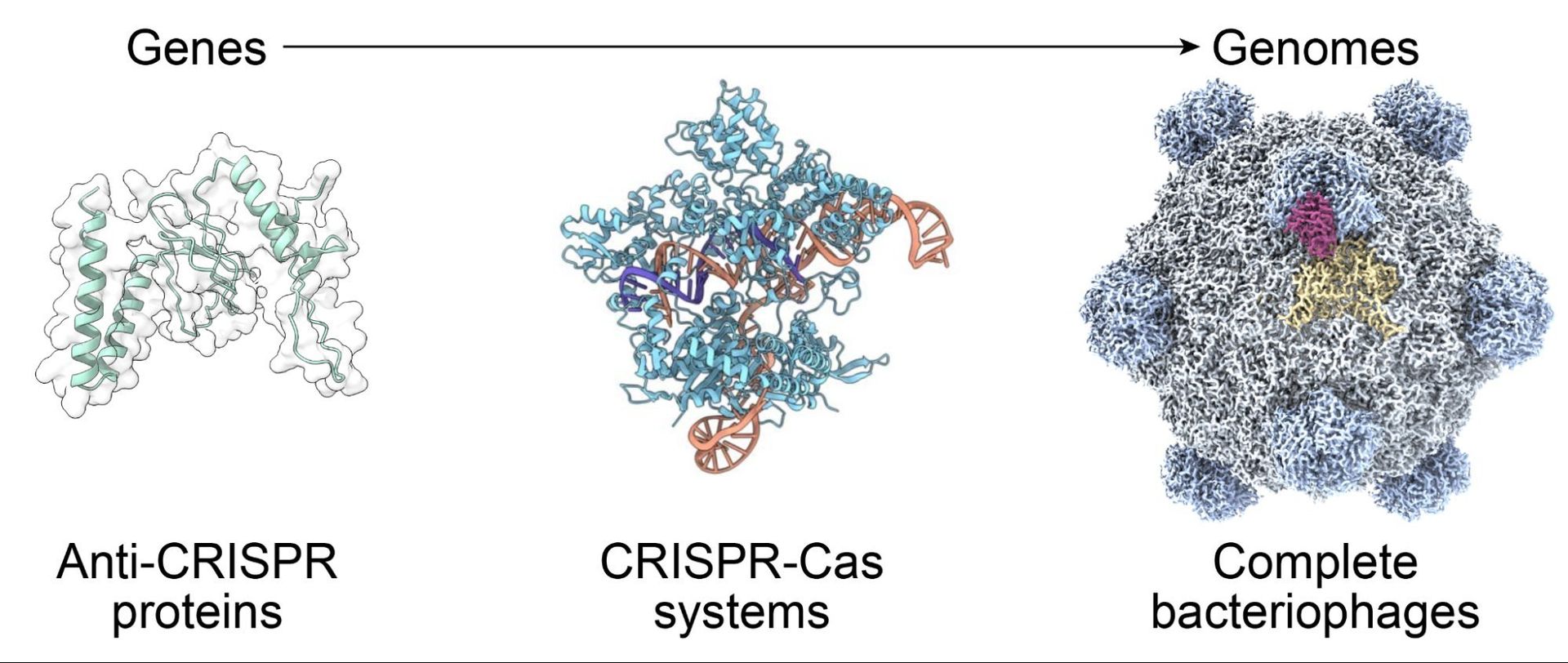
For decades, biologists have progressed from sequencing DNA to synthesizing it—but designing entire functional genomes has remained an elusive frontier. Now, a team at Arc Institute, led by Samuel King and Brian Hie, has crossed that threshold by leveraging AI to generate the first synthetic bacteriophage genomes. Published in a BioRxiv preprint, their work demonstrates how genomic foundation models can orchestrate complex biological systems, opening transformative possibilities for biotechnology and medicine.
The Genome Design Challenge
Designing a genome isn't just stitching genes together—it demands harmonizing overlapping reading frames, regulatory elements, and evolutionary fitness across thousands of nucleotides. As King and Hie note, "Going from designing individual genes to complete genomes is an incredibly challenging problem." 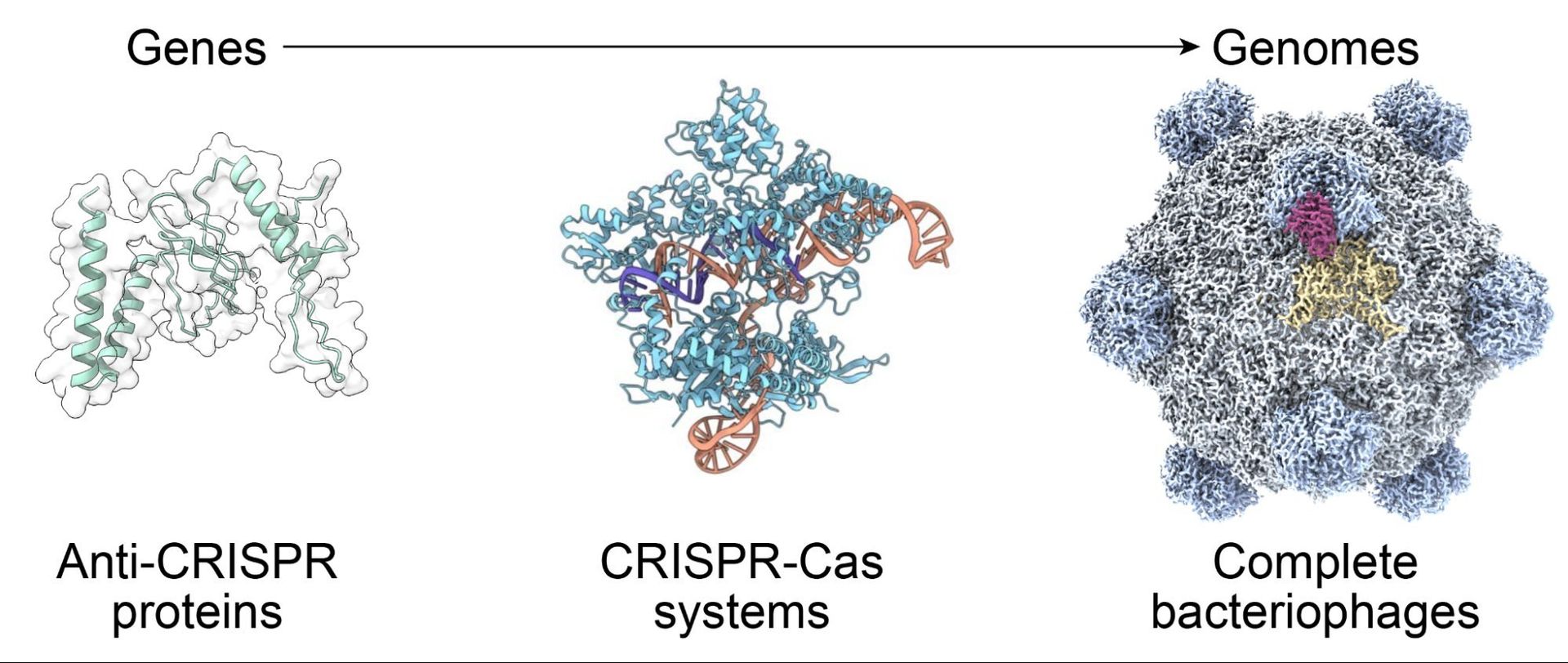 illustrates this complexity, where standard tools failed to identify all 11 genes in bacteriophage ΦX174 due to overlapping sequences. The team built a custom annotation pipeline combining ORF-finding and homology searches, which became critical for evaluating AI-generated designs.
illustrates this complexity, where standard tools failed to identify all 11 genes in bacteriophage ΦX174 due to overlapping sequences. The team built a custom annotation pipeline combining ORF-finding and homology searches, which became critical for evaluating AI-generated designs.
Why ΦX174? A Template for Making History
ΦX174, the first genome ever sequenced (1977) and synthesized (2003), was chosen for its balance of complexity and practicality. At 5,386 nucleotides, it encodes 11 genes with intricate overlaps, creating a "stringent test case" where mutations must satisfy multiple proteins simultaneously. As Hie explains, "This progression represents the fundamental capabilities that define modern genomics: we learned to read DNA, then to write it, and now to design it." 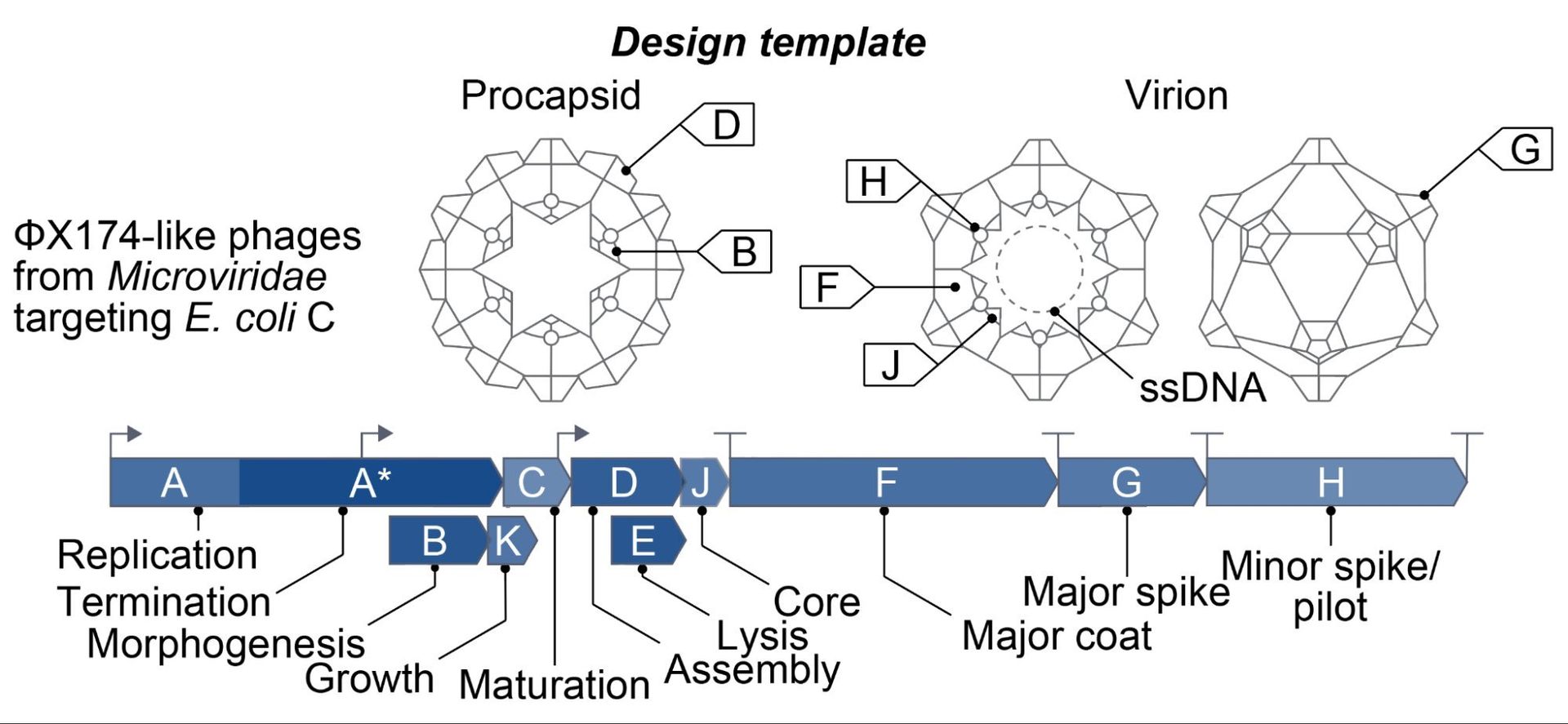 underscores how AI now drives this third revolution.
underscores how AI now drives this third revolution.
Engineering Evo: Fine-Tuning for Precision
The researchers fine-tuned their Evo genomic foundation model—pre-trained on millions of phage sequences—on a curated dataset of 14,466 Microviridae genomes. This specialization enabled controlled generation of ΦX174-like sequences through prompt engineering. "Careful tuning of the prompt and sampling parameters was essential," the paper details, ensuring novelty without straying from functional viability. Generated genomes were filtered for host specificity (targeting E. coli) and gene preservation, with only 16 of 285 designs proving functional after rigorous screening.
Validating AI’s Evolutionary Creativity
Experimental validation revealed AI’s capacity to innovate beyond nature. Functional genomes harbored 67–392 novel mutations, with one candidate (Evo-Φ2147) differing enough to qualify as a new species. Cryo-EM analysis confirmed that Evo-Φ36 successfully incorporated a protein from distantly related phage G4—a feat previously unattainable through rational design. Most strikingly, AI-generated phages overcame antibiotic-resistant E. coli strains in 1-5 passages, while natural ΦX174 failed. Mosaic genomes from multiple AI designs recombined to target bacterial receptors, proving that synthetic diversity can outpace resistance.
"The AI’s exploration of sequence space provides raw material for rapid adaptation," the authors write. "This transforms phage therapy from a trial-and-error process into a systematic approach for staying ahead of bacterial evolution."
From Phages to a New Genomic Era
Near-term applications include targeting pathogens like Pseudomonas aeruginosa, but the implications run deeper. As synthesis costs fall and models improve, whole-genome design could unlock biological possibilities never sampled by natural evolution. This work isn’t just a technical milestone—it redefines our relationship with biology, turning genomes into programmable substrates for innovation in therapeutics, agriculture, and beyond.
Comments
Please log in or register to join the discussion