NVIDIA’s transition from rendering video‑game explosions to decoding the language of life illustrates how GPU‑accelerated AI can solve one of biology’s longest‑standing puzzles—protein folding—and unlock a new era of precision drug design. The story blends deep science with cutting‑edge engineering, showing why a graphics card now powers the next generation of therapeutics.
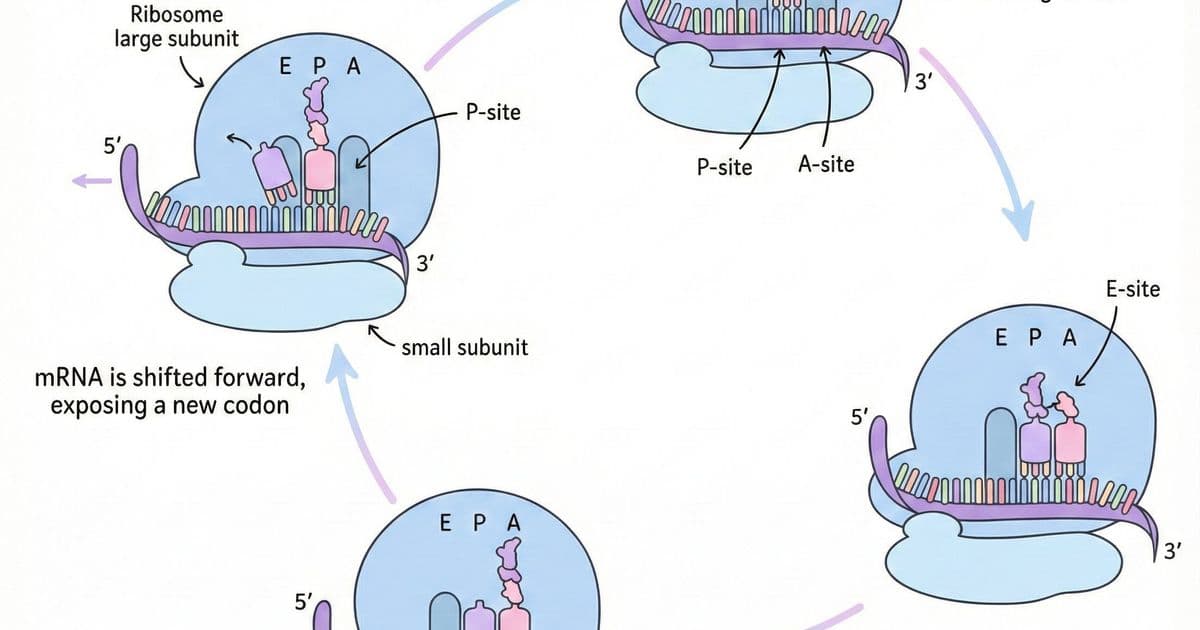
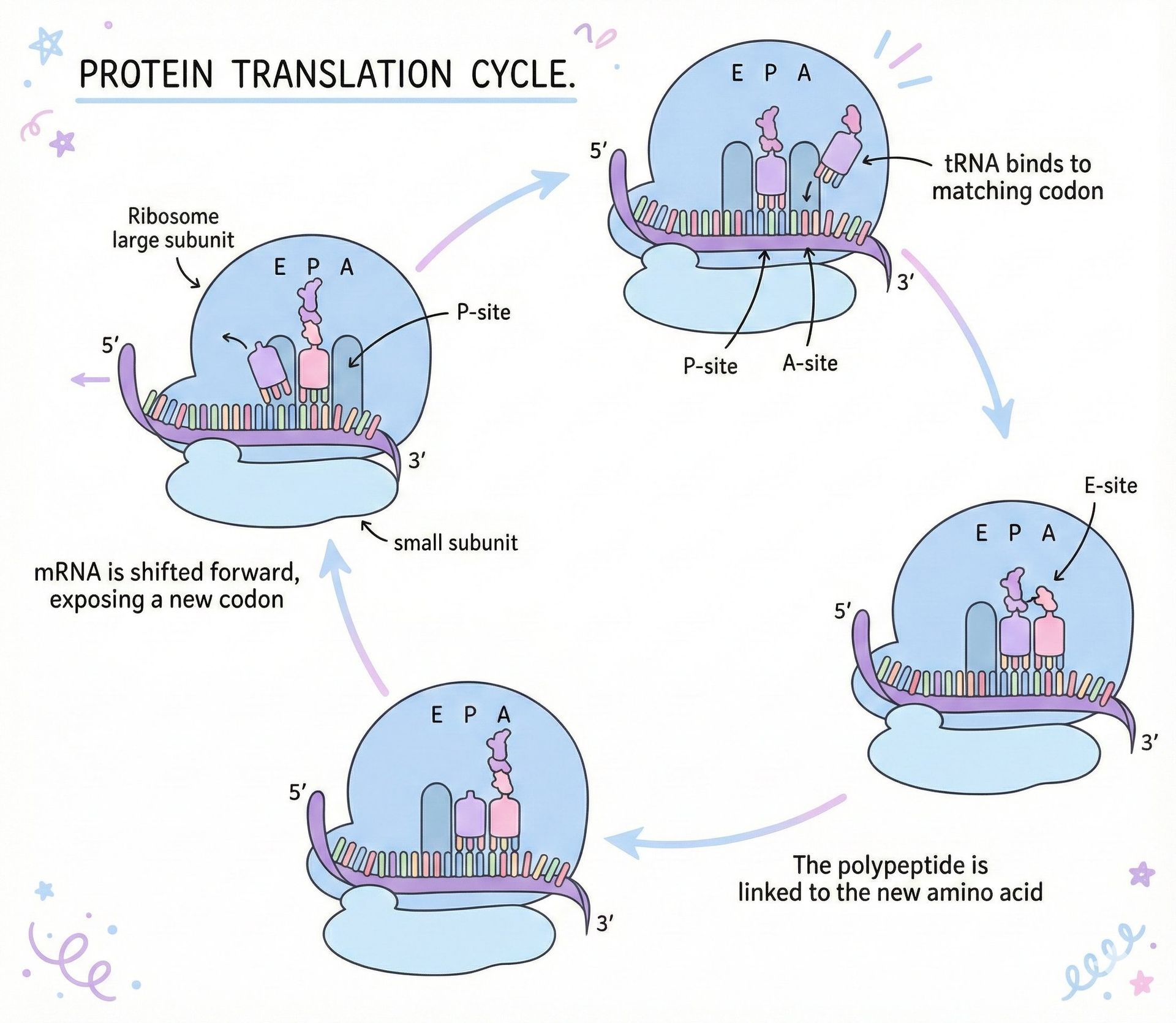 \n\nThe last decade has seen a quiet revolution in biology: a game‑chip that once powered the latest consoles is now the engine behind life‑saving medicines. NVIDIA’s GPUs, prized for rendering realistic graphics, have become the workhorse of deep learning models that predict, design, and even synthesize proteins—tiny machines that perform the chemistry of life.\n\n## The Protein Folding Problem\n\nAt its heart, a protein is a linear chain of 20 standard amino acids. When the ribosome finishes translating the chain, the polypeptide spontaneously folds into a three‑dimensional shape that determines its function. Misfolding can cause Alzheimer’s, cystic fibrosis, and prion diseases.\n\nThe search space for possible folds is astronomically large. A 100‑residue chain has roughly 10^95 potential conformations, yet proteins fold in milliseconds. This paradox, first highlighted by Cyrus Levinthal in 1969, underscores that folding is not a random walk but a guided descent down an energy funnel shaped by chemical interactions—hydrophobic collapse, hydrogen bonds, salt bridges, disulfide staples, and van der Waals forces.\n\n> “The native structure is the functional form; even a single amino‑acid change can render a protein inert or pathogenic.”\n\n## Why Accurate Structure Prediction Matters\n\n1. Disease Insight – Knowing a protein’s shape reveals how mutations disrupt function.\n2. Drug Design – Structural knowledge enables rational binding‑site targeting.\n3. Synthetic Biology – Designing enzymes or novel proteins opens new metabolic pathways.\n4. Evolutionary Biology – Comparing structures across species illuminates functional constraints.\n\n## AI Breakthroughs: AlphaFold 1 and 2\n\nDeepMind’s AlphaFold leveraged deep learning to learn patterns from over 170,000 experimentally determined protein structures. AlphaFold 1 (2018) achieved a median GDT score of 58.9, a modest improvement over traditional methods. AlphaFold 2 (2020) shattered expectations with a median GDT of 92.4, surpassing experimental accuracy for 87% of CASP14 targets.\n\nThe key innovation was treating the protein sequence like a language: the model learned attention patterns that implicitly encode spatial relationships, bypassing the need for explicit physics simulation.\n\n## NVIDIA’s GPU‑Powered Ecosystem\n\nNVIDIA’s GPUs excel at parallel matrix operations, making them ideal for training and running transformer‑based models. The company extended its hardware advantage into biology with several initiatives:\n\n- BioNeMo – A family of protein language models that generate embeddings and predict properties directly from sequences.\n- ProteinDT – A design platform where users describe desired functions in plain English, and the AI proposes amino‑acid sequences that meet those specifications.\n- La‑Proteina – Generates entirely novel folds not present in current databases, enabling creation of proteins that could, for example, degrade plastic or sequester carbon.\n- OpenFold – An open‑source implementation of AlphaFold optimized for NVIDIA GPUs, achieving up to 138× speedups.\n\nThese tools are already in the pipelines of pharma giants such as Pfizer, Amgen, and AstraZeneca, accelerating the discovery of antibody therapeutics, enzyme replacements, and small‑molecule inhibitors.\n\n## The Science Behind the Speed\n\nWhile AlphaFold 2 delivers near‑experimental accuracy, each prediction can take hours on a single GPU. NVIDIA’s optimizations reduce runtime by streamlining tensor operations, exploiting mixed‑precision arithmetic, and parallelizing over multiple GPUs in a cluster. The result is a scalable platform where thousands of candidate proteins can be screened in days rather than months.\n\n## From Protein to Product\n\nA typical drug‑discovery workflow now looks like this:\n\n1. Define the Target – Identify a protein implicated in disease and the desired functional outcome.\n2. Generate Candidates – Use ProteinDT or La‑Proteina to produce sequences that fold into the target shape.\n3. Validate Structure – Run OpenFold or BioNeMo to confirm the predicted 3D conformation and assess stability.\n4. Test Binding – Employ docking simulations and in‑vitro assays to measure affinity for the therapeutic target.\n5. Iterate – Refine sequences based on feedback, leveraging GPU‑accelerated training for rapid convergence.\n\nBecause the entire pipeline is computational, the bottleneck shifts from wet‑lab synthesis to in‑silico iteration, dramatically shortening the time from idea to clinical candidate.\n\n## The Broader Impact\n\nBeyond drug discovery, NVIDIA’s protein AI is already influencing environmental science—designing enzymes that break down microplastics—and agriculture—engineering crops with enhanced nutrient profiles. The convergence of GPU power, transformer models, and protein biology heralds a future where synthetic organisms can be tailored for specific societal challenges.\n\n> “We are no longer waiting for evolution to find a solution; we can now design the next generation of proteins in a matter of hours.”\n\n## Conclusion\n\nThe journey from gaming GPUs to life‑saving molecules exemplifies how cross‑disciplinary innovation can solve problems once thought intractable. NVIDIA’s hardware, coupled with transformer‑based AI, has turned the protein folding problem from a 50‑year puzzle into a tractable, industrial‑scale workflow. As these tools mature, the next wave of therapeutics will be engineered, not discovered, marking a paradigm shift in biomedical research.\n\nSource:
\n\nThe last decade has seen a quiet revolution in biology: a game‑chip that once powered the latest consoles is now the engine behind life‑saving medicines. NVIDIA’s GPUs, prized for rendering realistic graphics, have become the workhorse of deep learning models that predict, design, and even synthesize proteins—tiny machines that perform the chemistry of life.\n\n## The Protein Folding Problem\n\nAt its heart, a protein is a linear chain of 20 standard amino acids. When the ribosome finishes translating the chain, the polypeptide spontaneously folds into a three‑dimensional shape that determines its function. Misfolding can cause Alzheimer’s, cystic fibrosis, and prion diseases.\n\nThe search space for possible folds is astronomically large. A 100‑residue chain has roughly 10^95 potential conformations, yet proteins fold in milliseconds. This paradox, first highlighted by Cyrus Levinthal in 1969, underscores that folding is not a random walk but a guided descent down an energy funnel shaped by chemical interactions—hydrophobic collapse, hydrogen bonds, salt bridges, disulfide staples, and van der Waals forces.\n\n> “The native structure is the functional form; even a single amino‑acid change can render a protein inert or pathogenic.”\n\n## Why Accurate Structure Prediction Matters\n\n1. Disease Insight – Knowing a protein’s shape reveals how mutations disrupt function.\n2. Drug Design – Structural knowledge enables rational binding‑site targeting.\n3. Synthetic Biology – Designing enzymes or novel proteins opens new metabolic pathways.\n4. Evolutionary Biology – Comparing structures across species illuminates functional constraints.\n\n## AI Breakthroughs: AlphaFold 1 and 2\n\nDeepMind’s AlphaFold leveraged deep learning to learn patterns from over 170,000 experimentally determined protein structures. AlphaFold 1 (2018) achieved a median GDT score of 58.9, a modest improvement over traditional methods. AlphaFold 2 (2020) shattered expectations with a median GDT of 92.4, surpassing experimental accuracy for 87% of CASP14 targets.\n\nThe key innovation was treating the protein sequence like a language: the model learned attention patterns that implicitly encode spatial relationships, bypassing the need for explicit physics simulation.\n\n## NVIDIA’s GPU‑Powered Ecosystem\n\nNVIDIA’s GPUs excel at parallel matrix operations, making them ideal for training and running transformer‑based models. The company extended its hardware advantage into biology with several initiatives:\n\n- BioNeMo – A family of protein language models that generate embeddings and predict properties directly from sequences.\n- ProteinDT – A design platform where users describe desired functions in plain English, and the AI proposes amino‑acid sequences that meet those specifications.\n- La‑Proteina – Generates entirely novel folds not present in current databases, enabling creation of proteins that could, for example, degrade plastic or sequester carbon.\n- OpenFold – An open‑source implementation of AlphaFold optimized for NVIDIA GPUs, achieving up to 138× speedups.\n\nThese tools are already in the pipelines of pharma giants such as Pfizer, Amgen, and AstraZeneca, accelerating the discovery of antibody therapeutics, enzyme replacements, and small‑molecule inhibitors.\n\n## The Science Behind the Speed\n\nWhile AlphaFold 2 delivers near‑experimental accuracy, each prediction can take hours on a single GPU. NVIDIA’s optimizations reduce runtime by streamlining tensor operations, exploiting mixed‑precision arithmetic, and parallelizing over multiple GPUs in a cluster. The result is a scalable platform where thousands of candidate proteins can be screened in days rather than months.\n\n## From Protein to Product\n\nA typical drug‑discovery workflow now looks like this:\n\n1. Define the Target – Identify a protein implicated in disease and the desired functional outcome.\n2. Generate Candidates – Use ProteinDT or La‑Proteina to produce sequences that fold into the target shape.\n3. Validate Structure – Run OpenFold or BioNeMo to confirm the predicted 3D conformation and assess stability.\n4. Test Binding – Employ docking simulations and in‑vitro assays to measure affinity for the therapeutic target.\n5. Iterate – Refine sequences based on feedback, leveraging GPU‑accelerated training for rapid convergence.\n\nBecause the entire pipeline is computational, the bottleneck shifts from wet‑lab synthesis to in‑silico iteration, dramatically shortening the time from idea to clinical candidate.\n\n## The Broader Impact\n\nBeyond drug discovery, NVIDIA’s protein AI is already influencing environmental science—designing enzymes that break down microplastics—and agriculture—engineering crops with enhanced nutrient profiles. The convergence of GPU power, transformer models, and protein biology heralds a future where synthetic organisms can be tailored for specific societal challenges.\n\n> “We are no longer waiting for evolution to find a solution; we can now design the next generation of proteins in a matter of hours.”\n\n## Conclusion\n\nThe journey from gaming GPUs to life‑saving molecules exemplifies how cross‑disciplinary innovation can solve problems once thought intractable. NVIDIA’s hardware, coupled with transformer‑based AI, has turned the protein folding problem from a 50‑year puzzle into a tractable, industrial‑scale workflow. As these tools mature, the next wave of therapeutics will be engineered, not discovered, marking a paradigm shift in biomedical research.\n\nSource:
Comments
Please log in or register to join the discussion