Large language models have transformed knowledge work by predicting the next token. Now, the Arc Institute's Evo 2 applies this same principle to biological sequences, demonstrating remarkable accuracy in predicting genetic mutations with zero-shot learning and even generating functional genomic sequences.
Applying Language Model Architecture to Biology: Evo 2's Breakthrough in Genomic Prediction
Large language models have transformed knowledge work by predicting the next token. Now, the Arc Institute's Evo 2 applies this same principle to biological sequences, demonstrating remarkable accuracy in predicting genetic mutations with zero-shot learning and even generating functional genomic sequences.
The Challenge of Biological Sequences
While large language models have revolutionized how we process and generate human language, applying similar techniques to biological data presents unique challenges. Consider the human genome—a complex structure with 6 billion base pairs arranged in intricate hierarchical relationships. Unlike text where words follow relatively predictable patterns, biological sequences have long-range dependencies where elements millions or even billions of base pairs apart can interact physically and functionally.
 Evo 2's architecture addresses these challenges through a modified attention mechanism designed specifically for biological sequences.
Evo 2's architecture addresses these challenges through a modified attention mechanism designed specifically for biological sequences.
Evo 2: A Biological Foundation Model
The Arc Institute has introduced Evo 2, a groundbreaking biological foundation model that tackles these unique challenges in genomic data. Built on a modified attention architecture, Evo 2 features up to 40 billion parameters and an impressive 1-million-token context window—essential for capturing the long-range dependencies in genomic sequences.
Trained on over 9 trillion nucleotides, Evo 2 has demonstrated remarkable capabilities:
- Predicting the effects of genetic mutations with zero-shot accuracy
- Generating functional genomic sequences from scratch
- Understanding complex hierarchical relationships in DNA
Installing Evo 2: A Practical Guide
For developers and researchers looking to experiment with Evo 2, proper installation is crucial. As a deep learning model, Evo 2 performs best on Linux environments with Nvidia GPUs. Windows users should consider switching to WSL (Windows Subsystem for Linux) before installation.
Prerequisites
Install Conda: If you don't have Conda already, download miniconda:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh bash Miniconda3-latest-Linux-x86_64.shProceed with initialization and configure:
conda config --set auto_activate_base falseClone the Evo 2 Repository:
git clone https://github.com/arcinstitute/evo2 cd evo2Create and Activate Conda Environment:
conda create -n evo2 python=3.12 conda activate evo2Install Prerequisites:
conda install -c nvidia cuda-nvcc cuda-cudart-dev conda install -c conda-forge transformer-engine-torch=2.3.0 conda install psutil pip install flash-attn==2.8.0.post2 --no-build-isolation pip install -e .Install GCC (if needed):
which gcc # Check if gcc is installed sudo apt-get update sudo apt-get install build-essential # Install if neededVerify Installation:
python -m evo2.test.test_evo2_generation --model_name evo2_7bThe script should complete with a PASS.
Case Study: BRCA1 Mutation Prediction
A compelling first application of Evo 2 is predicting the effects of mutations in the BRCA1 gene (BReast CAncer gene 1). This gene encodes a protein crucial for repairing damaged DNA, and mutations can significantly increase the risk of breast or ovarian cancers.
Zero-shot Learning Approach
Evo 2 can predict mutation effects without specific training on BRCA1 data—a capability known as zero-shot learning. The model calculates how "normal" a mutated sequence appears based on patterns learned during training on vast genomic datasets.
 The probability calculation demonstrates how Evo 2 evaluates sequence likelihood.
The probability calculation demonstrates how Evo 2 evaluates sequence likelihood.
Probability Calculations Explained
As an autoregressive model similar to ChatGPT, Evo 2 predicts the probability of each subsequent base in a sequence. For example, to calculate the probability of the sequence 'TACG':
The model first processes the beginning of sequence token
<BOS>and outputs probabilities for the next token. If 'T' has a 20% probability, then P(T) = 20%.With
<BOS>Tas input, the model predicts the next token. If 'A' has a 15% probability, then P(A|T) = 15%.With
<BOS>TAas input, the model predicts the next token. If 'C' has a 25% probability, then P(C|TA) = 25%.Finally, with
<BOS>TACas input, the model predicts the next token. If 'G' has a 20% probability, then P(G|TAC) = 20%.
The overall probability is the product of these conditional probabilities: P(TACG) = P(T) × P(A|T) × P(C|TA) × P(G|TAC) = 20% × 15% × 25% × 20% = 0.15%
Mutation Scores and Classification
The model calculates an Evo 2 mutation (delta) score as P(mutated sequence) – P(original sequence). A lower score indicates that the mutation disrupts the natural pattern of the sequence, suggesting a potential loss-of-function mutation.
Experimental data classifies mutations as either benign (FUNC/INT) or causing loss of function (LOF). When comparing model predictions against these classifications, benign mutations typically show higher delta scores, as they're more likely to appear naturally.
Improving Predictions: Model Size and Embeddings
Using the 7B Model
The initial BRCA1 analysis used the Evo 2 Base model with 1 billion parameters. Switching to the larger 7B model significantly improved performance:
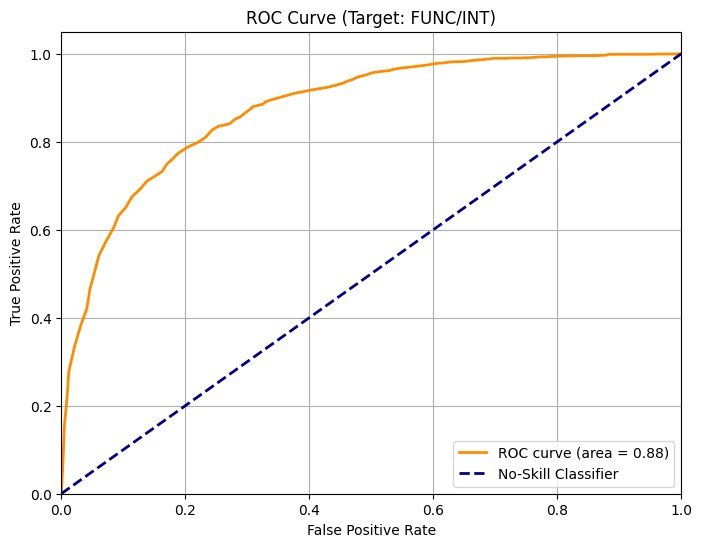
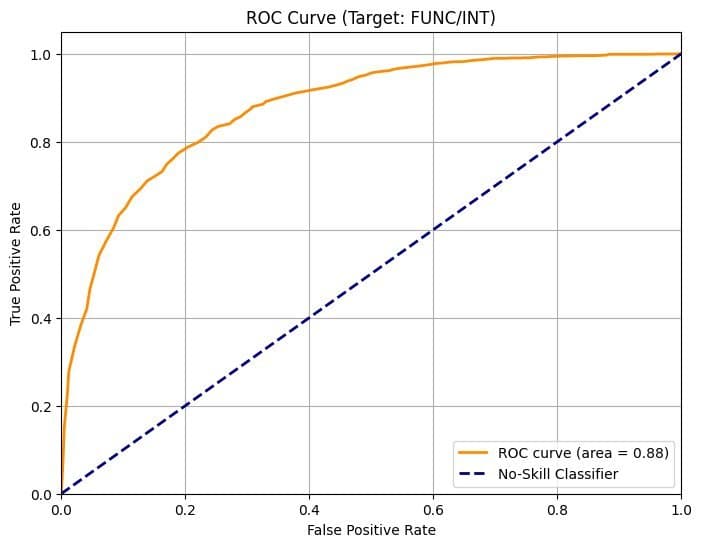
- Evo 2 1B: ROC AUC = 0.73
- Evo 2 7B: ROC AUC = 0.88
The larger model's distributions for benign and loss-of-function mutations became more separable, enhancing classification accuracy.
Few-shot Learning with Embeddings
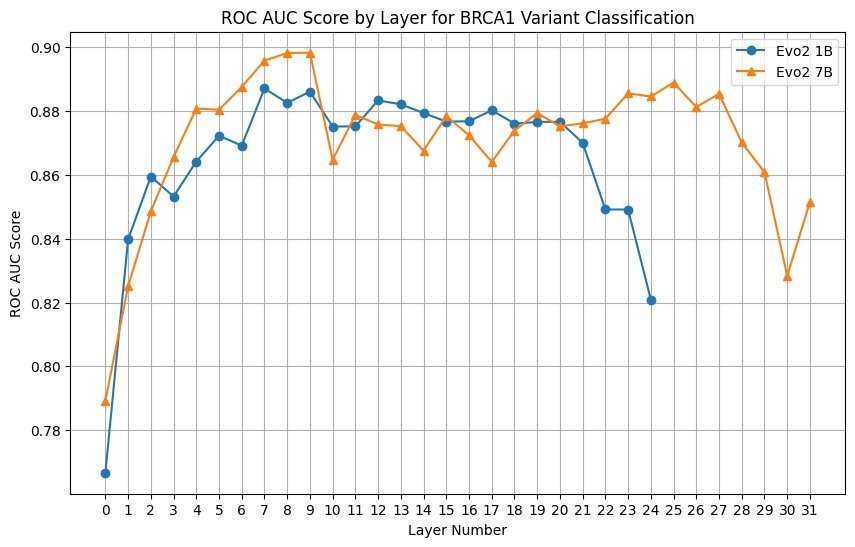
For even better performance, researchers can employ few-shot learning by training a classifier on some labeled data. This requires accessing sequence embeddings from the model's intermediate layers.
A layer sweep analysis revealed that, contrary to common wisdom, the best performing embeddings came from earlier layers rather than middle or later layers. Using embeddings from layer 20 with the 1B model improved the ROC AUC from 0.35 (zero-shot) to 0.88.
Layer Sweep Analysis
Testing all layers with both 1B and 7B models showed stable performance across layers, as long as they weren't among the first or last layers. This suggests that embeddings from various layers can be effective for genomic classification tasks.
The Future of AI in Genomic Analysis
Evo 2 represents a significant step forward in applying AI to biological problems. By adapting language model architectures to handle the unique challenges of genomic data, researchers can now predict mutation effects with remarkable accuracy—even without specific training on the target genes.
The ability to perform zero-shot mutation prediction opens new possibilities for analyzing genetic variants of unknown significance. As models like Evo 2 continue to evolve and incorporate more biological knowledge, we can expect even more accurate predictions of how genetic changes affect health and disease.
For developers and bioinformaticians, Evo 2 provides both a powerful tool for current research and a foundation for building more advanced biological AI systems in the future. The combination of large-scale training data, modified attention architectures, and innovative evaluation methods positions models like Evo 2 at the forefront of computational biology.
Source: This article is based on content from PredictBio Labs
Comments
Please log in or register to join the discussion