A team of researchers has built the first high‑level language interpreter that runs entirely within Conway’s Game of Life, demonstrating Turing completeness in a cellular automaton. By weaving a Lisp runtime into a custom 8‑state VarLife rule and then translating it to the classic Life pattern, the project showcases how complex software can emerge from simple rules, offering fresh insights for researchers and hobbyists alike.
Introduction
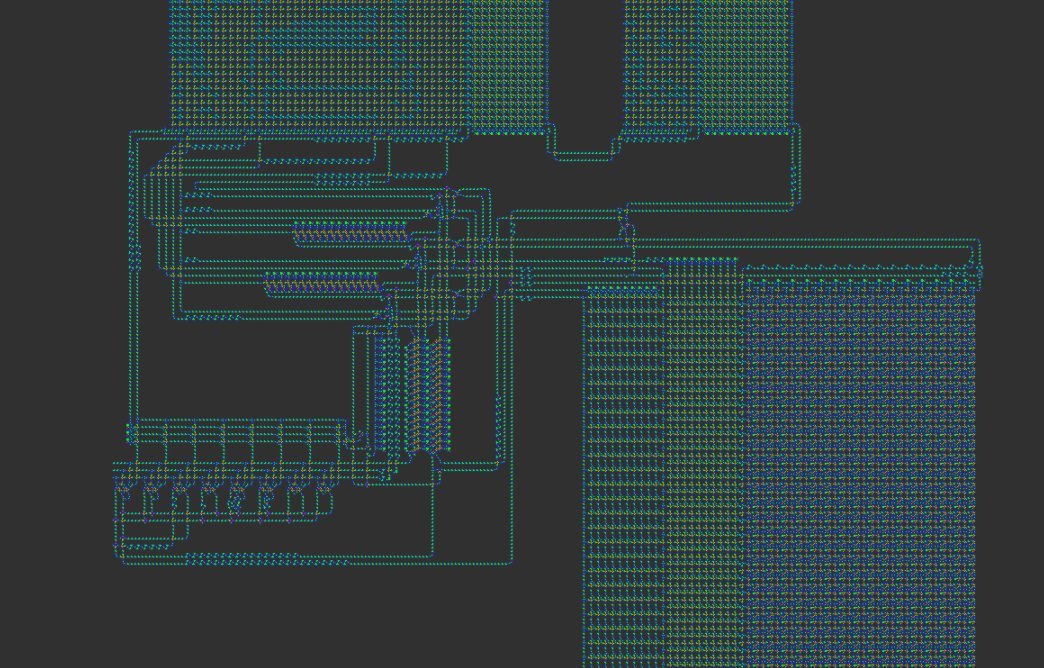
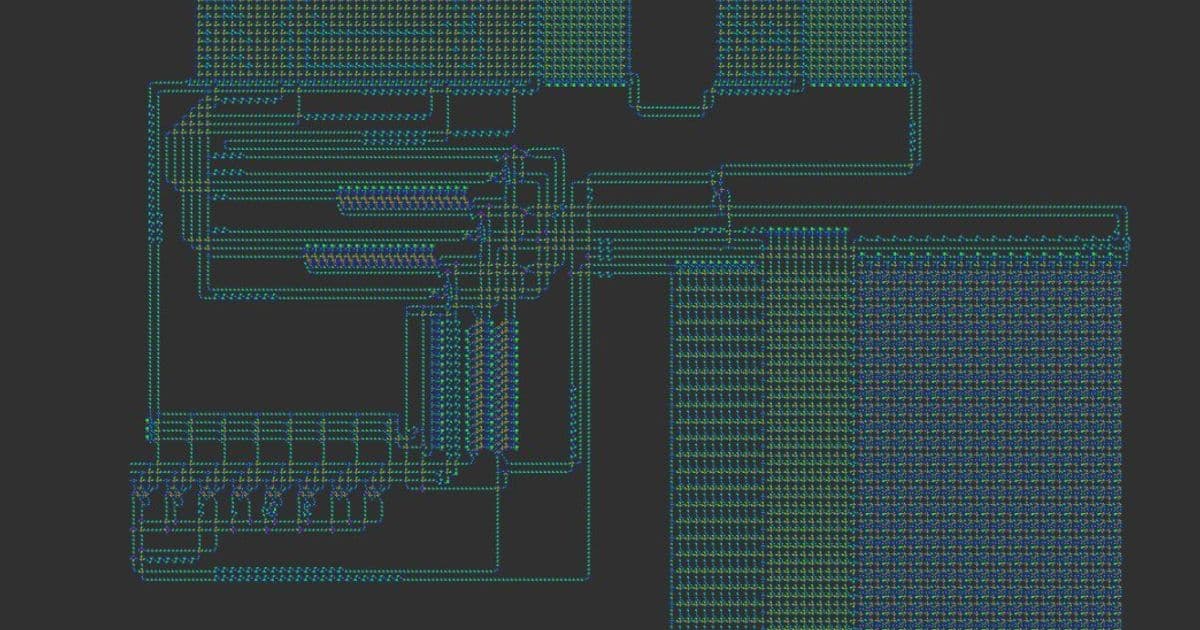
Lisp in Life is a groundbreaking project that embeds a complete Lisp interpreter into Conway’s Game of Life. The system was designed to prove that a high‑level language can be executed by a purely cellular automaton, a feat that had never been achieved before. The interpreter is written in C, compiled to an esoteric assembly language, and then mapped onto a custom 8‑state cellular automaton called VarLife. Finally, a conversion step turns the VarLife pattern into a standard Life pattern by using OTCA metapixels—tiny Life sub‑patterns that emulate arbitrary cellular rules.
From Lisp to Life: The Architecture
The architecture can be visualised in three layers:
- Lisp Runtime – A minimal interpreter that supports lexical closures, macros, and a small set of built‑in forms. It is written in C and heavily optimised for the target machine.
- QFT‑Inspired CPU – A custom CPU architecture originally defined by the Quest For Tetris (QFT) project. The CPU uses a ROM/RAM pair, a small register file, and a handful of opcodes. The interpreter is compiled to this architecture via an extended ELVM backend.
- VarLife → Life Conversion – The QFT machine is first instantiated in an 8‑state rule called VarLife. Each VarLife cell is represented by an OTCA metapixel in Conway’s Life, allowing the Life pattern to emulate the VarLife behaviour.
The overall design is illustrated in the screenshot below.
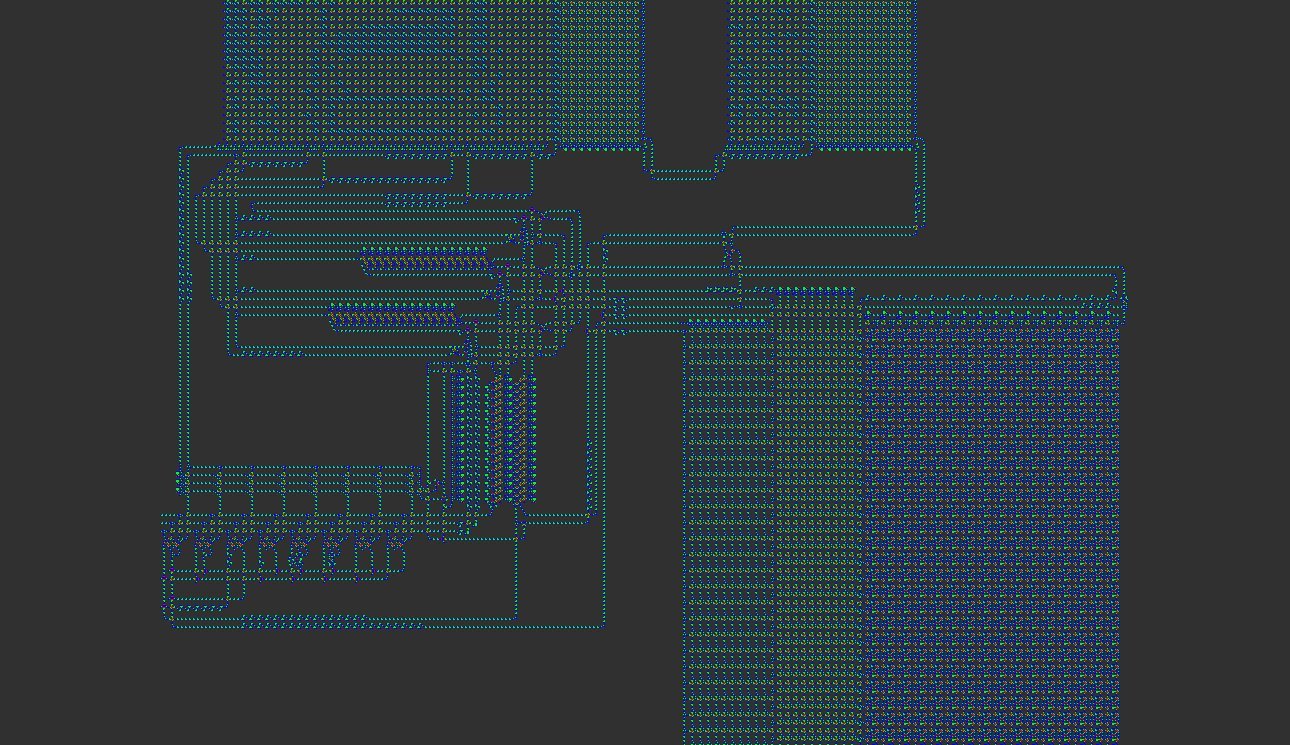
OTCA Metapixels and the VarLife Rule
OTCA metapixels are 2048×2048 Life sub‑patterns that carry their own birth/survival rules. By encoding each VarLife cell as a distinct metapixel, the conversion preserves the exact dynamics of the 8‑state rule while keeping the pattern compatible with standard Life viewers.
Optimisations that Made It Feasible
Running a Lisp interpreter in Life would normally be astronomically slow. The team introduced a cascade of optimisations across every layer:
- String Interning & Hash Tables – During parsing, symbols are stored in a 4‑bit hash table that points to a binary search tree. At evaluation time, symbol comparison reduces to integer equality.
- Jump‑Hash for Special Forms – 17 Lisp special forms are mapped to a hash table that allows constant‑time dispatch.
- Instruction Compression – The ROM is split into a lookup table and a value module, reducing the instruction word from 58 to 45 bits.
- Extended Address Space – ROM and RAM addresses grew from 9/7 bits to 12/10 bits, enabling larger programs.
- Macrocell and Hashlife Compression – The final Life pattern is stored in a quadtree format and run with Golly’s Hashlife algorithm, dramatically reducing simulation time.
These optimisations lowered the number of VarLife generations required for a simple program to just over 105 million, and the corresponding Life pattern to roughly 3.7 trillion generations—still a long run, but within the realm of practical simulation.
Performance Snapshot
| Program | VarLife Generations | Life Generations | VarLife Time | Life Time |
|---|---|---|---|---|
| print.lisp | 105 413 068 | 3 724 032 866 304 | 1.16 min | 382 min |
| factorial.lisp | 1 000 000 000 | 35 328 000 000 000 | 5.20 min | 3395 min |
While the Life pattern runs orders of magnitude slower, the relative scaling is consistent, confirming that the translation preserves the computational complexity of the underlying Lisp program.
Why It Matters
- Proof of Concept – The project is the first to demonstrate that a full high‑level language can be interpreted inside Life, reinforcing the theory that Life is Turing complete.
- Educational Tool – Students of computation theory can now experiment with a real interpreter that runs on a 2‑state cellular automaton, bridging abstract theory and hands‑on coding.
- Research Catalyst – The optimisation techniques—especially the OTCA metapixel conversion and instruction compression—may inspire new approaches to embedding virtual machines in unconventional substrates.
Looking Forward
The Lisp in Life architecture opens the door to more ambitious projects: a full compiler for a language, a more efficient virtual machine, or even a distributed Life‑based network. Each step will deepen our understanding of how complex software can be distilled into simple, local rules.
For those interested in experimenting, the source code and pattern files are publicly available:
- Repository: https://github.com/woodrush/lisp-in-life
- Live pattern viewer: https://woodrush.github.io/blog/posts/2022-01-12-lisp-in-life.html
A short video demonstrates a running instance of the interpreter.
The project showcases a remarkable blend of theoretical computer science, low‑level systems engineering, and artistic expression, proving that even the most abstract ideas can find a home in the humble grid of Life.
Source: https://woodrush.github.io/blog/posts/2022-01-12-lisp-in-life.html
Comments
Please log in or register to join the discussion